![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
95 Cards in this Set
- Front
- Back
|
1. Nucleic acids are polymers:polynucleotides
2. Monomeric units: nucleotides 3. Nucleotides are joined together by 3', 5' phosphodiester bonds |
Basic concepts of nucleic acid structure
|
|
|
Major Purine bases: Adenine and guanine
Major pyrimidine bases: thymine and cytosine Sugar moiety: Deoxyribose |
DNA
|
|
|
Major Purine bases: Adenine and guanine
Major pyrimidine bases: uracil and cytosine Sugar moiety: Ribose |
RNA
Can have trace amounts of thymine. |
|
|
In modified bases of nucleic acids, _______________ occur at the level of polynucleotide
|
Modifications
Must incorporate the parent first. All have a function in the nucleic acid |
|
|
1. Polymers
2. Monomeric unit: nucleotide 3. Sugar-phosphate groups form the backbone 4. Bases are free to interact with bases in another strand 5. Phosphates carry a negative charge (polyanionic) 6. Directionality |
Structure and properties of polynucleotides
By convention polynucleotides are written in the 5’ to 3’ direction from left to right. |
|
|
If X is OH, then the sugar is ribose and the structure is _____
|
RNA
|
|
|
If X is H, then the sugar is
deoxyribose and the structure is _______ |
DNA
|
|
|
1. Sanger technique
i. Most common/Preferred. ii. Isolate DNA, ship it off in mail and wait. 2. Maxum- Gilbert |
Two techniques for sequencing DNA
|
|
|
1. Primary: sequence of nucleotides
2. Secondary: double-stranded DNA duplex 3. Tertiary: superhelicity |
DNA Structure
|
|
|
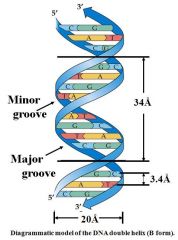
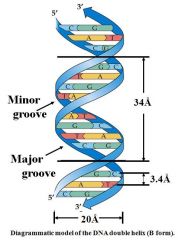
1. Antiparallel orientation of strands
2. Bases packed in interior of helix 3. Phosphate & sugar groups form backbone of two strands 4. Two grooves (major & minor) extending the length of the duplex |
Properties of a double-stranded helix
|
|
|
Distance between adjacent base-pairs
About 10 base pairs for B form of helix |
Rise
|
|
|
Distance between adjacent base-pairs.
|
Pitch
|
|
|
Hydrophobic
Packed into duplex away from aqueous environment No water molecules between base pairs |
Bases
|
|
|
Hydrophilic outside
|
Sugar/phosphate backbone
|
|
|
Function:
Proteins recognize sequence by binding into major grooves. Histones stabilize minor grooves |
Grooves
|
|
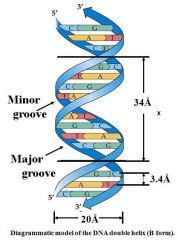
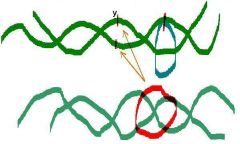
X is the distance for one helical turn
|
Pitch
|
|
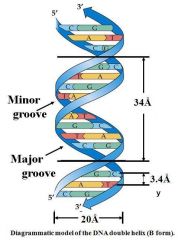
y is the distance between adjacent base-pairs
|
Rise
|
|
|
_________ can bind to DNA without disrupting the H-bonding between the two bases.
|
Protein
|
|
|
Proteins that bind to DNA with specificity recognize the respective bases through interactions in the _____or _______ groove.
|
major or minor groove
|
|
|
Most proteins will bind in _______ groove
More space for binding |
major
|
|
|
Name the groove:
Histones interact with DNA |
Minor groove
|
|
|
10.4
|
The number of base pairs for B-form
A form has 11 and Z form has 12. |
|
|
Predominant form of DNA is ___-form
|
B
|
|
|
____ form only found during replication
|
A
|
|
|
___-form requires certain sequence of DNA in order to form.
This form of DNA is found in regions containing alternating GC base-pairs in which the cytosine bases are methylated. |
Z
|
|
|
___-form is more elongated than B-form
Has a smaller diameter Major groove much longer Almost no minor groove. |
Z
|
|
|
___ form is more compact, with a larger diameter
Both have similar sized major grooves |
A
|
|
|
1. Hydrogen bonding
2. Base stacking: A. Hydrophobic interactions B. Van der Waals interactions(induced dipole moments) |
The two major forces that stabilize double stranded DNA complex.
Guanine-cytosine complex is more stable (3 hydrogen bonds vs. 2) |
|
|
1. Watson-crick base pairing
2. Complementary 3. Chargaff's rule |
Stability concepts
|
|
|
i. A binds to T
ii. C binds to G |
2. Complementary
|
|
|
i. Native double stranded DNA only
ii. The amount of A has to equal the amount of T (likewise for C and G) |
3. Chargaff's rule
|
|
|
i. Hydrogen bonds that form between A-T and C-G
|
1. Watson-crick base pairing
|
|
|
A. Increased Temperature
B. Decreased pH C. Increased pH D. Decreased Ionic strength (salt concentration) E. Chemical reagents : Urea, formamide etc. |
Conditions leading to denaturation
|
|
|
- DNA has net negative charge.
- Unstable - Repels the other strand - Stabilized by: Na+ K+ Mg2+ |
Why low salt concentration affects DNA
|
|
|
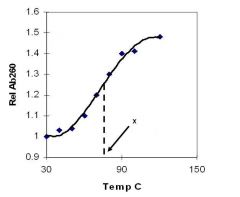
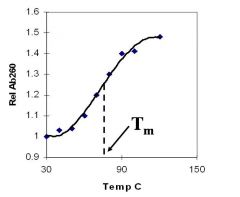
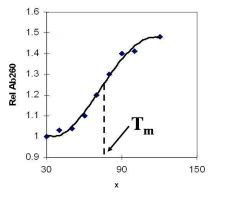
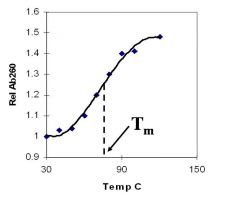
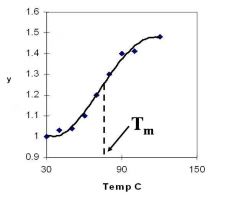
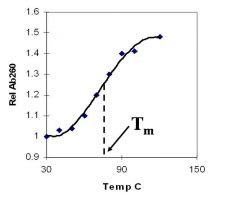
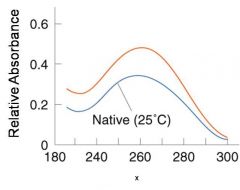
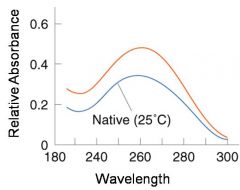
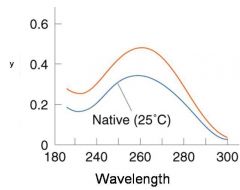
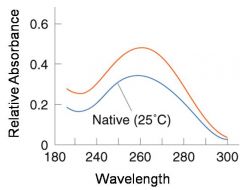
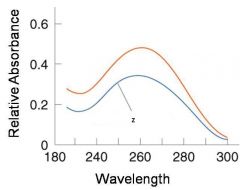
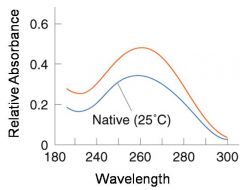
A double-stranded DNA duplex absorbs less light near 260 nm than the two strands that make it up do separately
When the molecules melt (come apart at higher temperatures), the absorbance increases The mid-point of the melting curve, when half of the bases no longer are hydrogen bonded to each other, is called Tm. |
The hyperchromic effect
|
|
|
Corresponds to mid point of curve
50% helical content Dependent on composition of DNA |
Tm = melting point of DNA
GC (120 C) has a higher melting point than AT (75 C) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1. Must align the two strands by interaction with bases
Complementary sequences must encounter one another and align properly. Can take hours to days 2. After alignment, rapid formation of the duplex. |
Process of Renaturation (reannealing)
|
|
|
Negative supercoiling - deficit of helical turns
Positive supercoiling - excess of helical turns |
Tertiary structure (superhelicity)
|
|
|
Deficit of helical turns
|
Negative supercoiling
A histone can fit inside the negative supercoiled group. Predominantly, negatively supercoiled form inside the cell. |
|
|
Excess of helical turns
|
Positive supercoiling
|
|
|
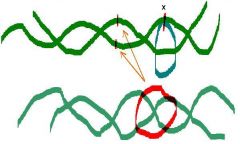
1. Type I topoisomerase activit (Transient cut of one strand)
2. Type II topoisomerase activity (Transient cut of both strands) |
Topoisomerases regulate the extent of supercoiling
|
|
|
|
|
|
|
|
|
|
|
|
|
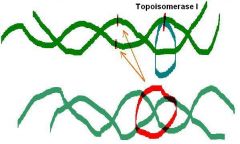
The ______ protein from E. coli (topoisomerase I) removes negative supercoiling from the circular ds-DNA
Does not require ATP Turns negatively supercoiled form into relaxed form. |
The omega protein
|
|
|
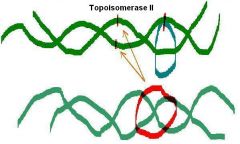
____ _______ from E. Coli (topoisomerase II)
Turns positively supercoiled form into relaxed form - Uses ATP Then turns relaxed form into negative supercoiled form - Uses ATP |
DNA gyrase
DNA gyrase Can remove positive supercoiling or introduce negative supercoiling. |
|
|
Through the action of:
1. DNA gyrase 2. The omega protein 3. Other topoisomerases The degree of supercoiling in DNA can be ________. |
regulated
|
|
|
ATP release energy is stored in the negatively supercoiled structure
Allows formation of the strained molecule Helical turns absent in the top region of duplex Have turns throughout the molecule |
The energy stored in the negatively supercoiled form can be used to disrupt base pairs under certain conditions.
This facilitates both replication and transcription. |
|
|
Topoisomerases can remove negative and positive supercoiling
Cannot induce negative supercoiling |
According to dogma, there are no topoisomerases that can directly introduce negative supercoiling into relaxed eukaryotic nuclear DNA.
|
|
|
Eukaryotic type I topoisomerase activity
|
Eukaryotic topoisomerase I and III
Negatively supercoiled form -> Relaxed form |
|
|
Eukaryotic type II topoisomerase activity
Positively supercoiled form + ATP -> Relaxed form Cannot isomerize relaxed form into negative supercoiled form. |
Eukaryotic topoisomerase II
|
|
|
To induce negative supercoiling, need binding of _______core.
|
histone
|
|
|
(a) Chromatin assembly on relaxed, closed-circular DNA.
(b) Binding of a histone core to form a nucleosome will induce one negative supercoil; but in the absence of any breaks, a positive supercoil must also form elsewhere in the DNA. (c) Relaxation of this positive supercoil by cellular topoisomerases leaves one net negative supercoil. |
Generation of a negative supercoiled core in eukaryotic DNA
Introduction of negative supercoiling in eukaryotic nuclear DNA is dependent upon interactions with histones |
|
|
1. Primary structure: sequence of nucleotides
2. Secondary structure: regions of double -stranded RNA 3. Tertiary structure: folding of the RNA in space |
RNA structure
Chargaff's rule: Single stranded RNA Not obeyed. [G] does not equal [C] [A] does not equal [U] Only in dsDNA is the rule obeyed. |
|
|
1. Double-stranded helical duplex; A-form geometry
2. Antiparallel orientation of strands 3. Phosphate & sugar groups form backbone of the two strands 4. Two grooves (major & minor) extending length of the duplex 5. Bases packed in interior of helix; A:U & G:C |
Properties of stem (secondary structure - regions of ds-RNA)
|
|
|
Biological function is dependent upon this RNA structure.
Has a cloverleaf secondary structure. Tertiary structure stabilized by T and D loops H-bonding |
tRNA
Has a well-defined secondary and tertiary structure. Contains a high number of modified bases |
|
|
Biological function is dependent upon this structure.
Four domains - Each domain will fold separately |
rRNA has a well-defined secondary and tertiary structure.
|
|
|
Secondary structure in _____ is thought to be involved in regulation of gene expression in certain cases.
Secondary structure differs from each type of mRNA Have numerous stems and loops Regulate gene formation at level of transcription and translation Hairpins can terminate translation, etc. |
mRNA
Overall, the structures of the three major forms of RNA dictate their function. |
|
|
Base catalyzed hydrolysis only acts on RNA, not DNA
DNA lacks the 2' hydroxyl group To isolate DNA from RNA, can treat the solution with base. |
Key point: Only RNA undergoes base-catalyzed hydrolysis.
|
|
|
1. Exonucleases
2. Endonucleases |
Types of enzymatic nucleases.
|
|
|
Cleave phosphodiester bonds located at either the 3' or 5' terminus
|
Exonucleases
|
|
|
Cleave phosphodiester bonds located in the interior
|
Endonucleases
|
|
|
a. Bacterial enzymes
b. Specificity - recognize palindromes Palindromes - inverted repeats |
Restriction endonucleases
Restriction enzymes cleave with specificity at palindromes. They serve an important role in the analysis of genomes. |
|
|
Bacteria that only infect bacteria
Inject DNA into host. |
Bacteriophages
|
|
|
Endonucleases are also located on bacteriophage DNA.
Why don't the endonucleases affect their DNA? |
Methylation at particular sites of base prevent endonuclease activity.
|
|
|
During replication, each strand has only part of the methyl groups.
|
Methylases only work on hemimethylated state
|
|
|
Does not have any methylation, cannot be methylated..
|
Viral DNA
|
|
|
1. Soak gel with a dye (ethidium bromide) that binds to DNA and fluoresces brightly under ultraviolet light .
2. Autoradiography: Prior to cleavage with restriction enzymes, label the DNA with the radioisotope 32P. - Since the beta particles emitted from 32P will expose photographic film, a sheet of film placed flat on the top of the agarose gel will show the position of DNA fragments when developed. 3. Other… |
Key concept: DNA fragments obtained from restriction enzyme digestion can be resolved by gel electrophoresis.
|
|
|
Southern blot analysis: DNA detection
Northern blot analysis: RNA detection (Western blot analysis: Protein detection) |
Analytical techniques
|
|
|
1. Separate a mixture of either DNA or RNA fragments by gel electrophoresis.
2. Transfer the nucleic acid fragments to a membrane such as nitrocellulose or PVDF (polyvinylidene difluoride) i.e. blotting. 3. Obtain a “probe” that is complementary to a specific region of the DNA or RNA. a. Soak the membrane in a solution containing the probe which may be labeled with 32P. b.The probe will hybridize to the nucleic acid fragment of interest. 4. Expose the membrane to X-ray film to visualize the probe “hybridized” to the nucleic acid of interest. |
Generic approach for DNA and RNA
|
|
|
Detection of:
1. Mutations in the human genome 2. Screening of food supply for pathogenes , 3. Testing for emerging infectious diseases |
Southern blotting
|
|
|
Determine differences in gene expression between:
- Tissues - Organs - Developmental stages - Environmental stress - Pathogen infection over the course of treatment |
Northern blotting:
|
|
|
Purpose:
1. Protects against damage 2. Reduces amount of space taken up by the DNA |
Packaging of DNA
|
|
|
1. Double-stranded circular DNA duplex
2. Negatively supercoiled 3. Neutralization of ionic charges on phosphate groups a. Cations: Na+, K+, Mg2+ others b. Polyamines |
Key concept: The prokaryotic genome is highly condensed
|
|
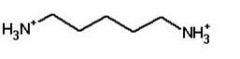
|
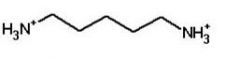
cadaverine
|
|
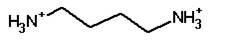
|
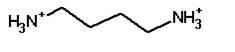
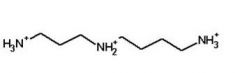
Putrescine
|
|
|
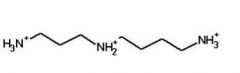
Spermidine
|
|
|
Packaging requires that you neutralize the ______ charges for negative supercoiling.
|
negative.
Use polyamines. |
|
|
Euchromatin
Loosely condensed and is the site of transcription. Heterochromatin Tightly condensed and NO transcription. |
Chromatin = DNA + Protein
Transcription is limited to regions of chromatin that are loosely condensed; i.e. euchromatin. |
|
|
DNA
1. Linear double-stranded duplexes 2. Negatively supercoiled Polyamines: putrescine, spermine & spermidine Cations: Na+, K+, Mg2+, others Proteins 1. Nonhistone proteins (Topoisomerases, DNA repair enzymes, RNA polymerase, etc.) 2. Histones |
Eukaryotic DNA Packaging
Eukaryotes do not have cadaverine Cannot be synthesized. |
|
|
Name the complex:
The H3-H4 tetramer interacts with the H2A-H2B dimer DNA is wrapped around the histone core. |
Octomeric complex
Histone octomer + DNA = Nucleosome |
|
|
Each histone has a tail that sticks through the DNA
Histones have a high arginine and lysine content |
Important for interaction with nucleosomes.
Allows for communication for different parts of DNA |
|
|
The chromatosome (two-turn
particle) consists of 166 bp of DNA (two superhelical turns). |
The H1 subunit is retained by this particle and may be associated with it.
|
|
|
The nucleosome consists of approximately ___ bp of DNA corresponding to 1 3/4 superhelical turns wound around a histone octamer.
|
146
|
|
|
Chromatosomes
containing less than 166 bp do not bind the ____ subunit. |
H1
|
|
|
In eukaryotic DNA, at each state, further condense DNA molecule, while maintaining functionality
Never will find naked DNA (DNA without histones) in nucleus unless you're dead |
Packaging of DNA in prokaryotes and eukaryotes differs in significant ways.
|
|
|
1. Most of the histones have been removed.
2. Protamines: proteins with a high content of Arg & Cys Polyamines Cations:Na+, K+, Mg2+, etc. |
In the sperm nucleus, protamines bind to the DNA, neutralizing its negative charge, and coiling the complex into tight circles .
These circles collapse into a "doughnut shaped structure.” The packaging of DNA in sperm is more compact than that in somatic cells. |
|
|
______ DNA in sperm (never occurs in nucleus)
- Interacts with protamine (lots of Arg and Cys) - Form dissulfide bonds between different protamines to form doughnut shape |
Naked
|
|
|
Required for neutralizing charge
|
Polyamines and cations
|

