![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
166 Cards in this Set
- Front
- Back
|
Alditol
|
Aldose reduced to an alcohol
|
|
|
Glycoside
|
Anomeric group on a saccharide reacting to become an alkoxy group on an acetal. Dehydration reaction.
|
|
|
Disaccharide formation and 3 examples
|
dehydration reaction between two monosaccharides
1. Sucrose: alpha - glucose - 1,2 - beta - fructose 2. Lactose: beta - galactose - 1,4 - beta - glucose 3. Maltose: alpha - glucose - 1,4 - beta - glucose |
|
|
Polysaccharide formation and 3 examples |
dehydration reaction between multiple monosaccharides. 1. cellulose: polymer of 1,4 - linked- beta glucose. (main structure in plants) 2. starch: polymer of linked - alpha glucose. Amylose is 1 - 4 linked alpha and amylopectin also has 1,6 links chaining off. Test for starch using iodine. Breakdown via alpha and beta amylase. 3. glycogen: similar to amylopectin, but with higher density of 1,6 linkages to allow for more sites for glycogen phosphorylase. |
|
|
Glycogen phosphorylase |
Cleaves glucose from nonreducing ends of glycogen |
|
|
alpha vs beta amylase |
alpha amylase randomly cleaves amylose to yield shorter polysaccharide chains, maltose, and glucose.
Beta amylase cleaves disaccharides off the nonreducing end of amylose to yield maltose. |
|
|
Amylose vs amylopectin vs glycogen |
Amylose is straight chain 1,4 alpha glucose polysaccharide chain. Amylopectin has more 1,6 branches. Glycogen has the most. Increased branching means more solubility and faster breakdown potential. |
|
|
Amphipathic molecules |
Containing both a hydrophobic and hydrophilic regions. |
|
|
Glycerophospholipids |
Head group ( variable) -- phosphodiester bond -- Glycerol backbone -- ester linkage -- 2X fatty acid tails All glycerophospholipids are phospholipids |
|
|
Sphingolipids. 4 major subclasses |
Sphingosine backbone. Not all contain phosphodiester linkages, so not all are classified as phospholipids. Instead some contain glycosidic linkages to sugars and are named glycolipids. Ceramides, sphingomyelins, glycolipids, gangliosides |
|
|
Glycolipid |
Sphingolipid with a glycosidic linkage to sugar instead of a phosphodiester bond. |
|
|
ceramide |
single hydrogen attached as head group of a sphingolipid. |
|
|
sphingomyelin |
a.k.a sphingophospholipids. These contain a phosphodiester bond with either a phosphatidylcholine or phosphatidylethanolamone as its head group no net charge on the head group major component in plasma membranes of myelin producing cells (oligodendrocytes and schwann cells) |
|
|
glycolipids, 3 types |
sphingolipids bound by glycosidic linkages. Not phospholipid. Mainly on outer surface of plasma membranes. May be further classified into cerebrosides (single sugar), globosides (two + sugar), and gangliosides. No net charge at physiologic pH for cerebrosides and globosides. Gangliosides have oligosaccharides with one or more N-acetylneuraminic acid (NANA or sialic acid) at the terminus and has a negative charge. Cell interaction, recognition, and signal transduction. |
|
|
Waxes |
Esters of long-chain fatty acids with long-chain alcohols (dehydration reaction)
RCO2H + ROH --> RCOOR + H2O |
|
|
Terpenes |
Isoprene (C5H8, 2-methylbuta-1,3-diene) are precursors. Monoterpene (10 carbons) = 2 isoprenes (2 x 5 carbons) sesquiterpene = 3 isoprenes diterpene = 4 isoprenes |
|
|
Steroids. What are they derived from? Describe the structure. |
Derived from terpenes. 3x cyclohexane + 1x cyclopentane base. Nonpolar and not necessarily always a hormone. |
|
|
Cholesterol. What type of hormone is it? What is the function? What is it a precursor to? |
A steroid hormone. Maintains membrane fluidity and is a precursor to many important molecules, including steroid hormones, bile acids, and vitamin K. Amphiphatic. |
|
|
Prostaglandin. What is it derived form? ____crine hormone? What does it regulate and how to inhibit it? |
Unsaturated carboxylic acids derived from arachidonic acid (20 carbons). Paracrine and autocrine hormones that often regulate production of cAMP. NSAIDS inhibit prostaglandin production by inhibiting COX. |
|
|
Fat soluble Vitamins (4) |
An essential nutrient that the body does not synthesize on its own. Vitamin A - Carotene. Unsaturated hydrocarbon important in vision, growth and development, and immune function. Aldehyde form is retinal, a component in the light sensing system in the eye. Retinol is oxidized to retinoic acid, which can regulate gene expression during epithelial development. Vitamin D - cholecalciferol. Activated to calcitriol in the liver and kidneys, which serve to increase calcium and phosphate intake in the intestines and promote bone production. Rickets is caused by vitamin D deficiency and results in underdeveloped, curved long bones and impeded growth. Vitamin E - tocopherols and tocotrienols. Substituted aromatic ring with a long isoprenoid side chain and hydrophobic. Biological antioxidants. Vitamin K - phylloquinone and menaquinones. Vital to post-translational modifications to form prothrombin. Also important for addition of calcium binding sites to many proteins. |
|
|
Triacylglycerides |
Three (usually different) fatty acid chains bound to glycerol via ester linkages. |
|
|
Free fatty acids |
Fatty acids not bound by ester linkages. They may circulate in blood attached noncovalently to serum albumin. |
|
|
Saponifaction |
Triacylglycerol + Strong base (KOH, NaOH) --> glycerol + free fatty acids (with cation counterion) |
|
|
Helicase |
Unravels DNA double helix |
|
|
Single Strand DNA binding protein |
Prevents unraveled DNA from annealing and from being denatured by nucleases |
|
|
DNA topoisomerase II |
A.k.a DNA gyrase relieves torsional strain during unraveling of DNA double helix by helicase. Introduces negative supercoils to offset the positive ones. |
|
|
DNA Polymerase. Reading direction and synthesizing direction. Prokaryotic vs eukaryotic versions |
Reads 3' to 5' on parent strand. Synthesizes daughter strand 5' to 3'. DNA Synthesis Prokaryotic: DNA polymerase III Eukaryotic: DNA polymerase alpha and delta Removal of RNA primers Prokaryotic: DNA polymerase I Eukaryotes: RNase H Replacement of RNA with DNA Prokaryotes: DNA polymerase I Eukaryotes: DNA polymerase delta |
|
|
Primase |
Adds ~10 nucleotide RNA primer to DNA for polymerase to latch on. |
|
|
DNA ligase |
Seals ends of okazaki fragments together. Lacks proofreading ability |
|
|
Eukaryotic DNA polymerases |
alpha and delta synthesize daughter strands. Delta fills in RNA primers with DNA Gamma replaces mitochondrial DNA beta and epsilon used in DNA repair delta and epsilon assisted by PCNA clamp (sliding clamp) |
|
|
Tumor suppressor genes 2 examples |
A.k.a antioncogenes P53 RB (retinoblastoma) Both must be silenced for loss of function |
|
|
Role of methylation in a DNA strand |
Allows for polymerase to determine which is the older parent strand (more methylated). Important during DNA repair since excised nucleoside should be on daughter strand. |
|
|
G2 phase DNA repair machinery |
Mismatch repair. In eukaryotes, repair enzymes encoded by MSH2, MLH1 In prokaryotes, MutS, MutL. |
|
|
Nucleotide excision repair. |
UV radiation induces thymine dimers, causing bulge in DNA backbone.
Excision endonuclease nicks backbone of damaged strand on either side of defective oligonucleotide and removes it. DNA polymerase resynthesizes correct sequence. Ligase finalizes repair. |
|
|
Base excision repair |
Glycosylase enzyme detects damaged bases and removes them, leaving behind an abasic site. AP endonuclease recognizes the site and removes damaged sequence from strand. DNA polymerase and ligase repair the removed section. |
|
|
Excision endonuclease |
Cuts out damaged thymine dimers in nucleotide excision repair |
|
|
Glycosylase |
Removes damages base, leaving an AP site behind |
|
|
AP endonuclease |
Removes damaged section in base excision repair. |
|
|
Recombinant DNA cloning via hosts |
1. Ligate DNA of interest, along with a gene for antibiotic resistance or other isolation technique, into a vector (virus or plasmid) 2. Infect host cells 3. Was colonies with antibiotic to select for ones where DNA of interest/antibiotic resistance gene has been transformed into genome 4. New bacteria colonies have DNA of interest in them. May use them to induce gene expression and make protein, or lyse and reacquire DNA. |
|
|
Restriction enzymes |
Endonucleases that cut at palindromic sites. May leave sticky ends. Allows for insertion of DNA of interest into vector DNA. |
|
|
cDNA |
Complementary DNA. Derived from mRNA, so includes only exons. Expression library. |
|
|
Southern blot. What does it find? Describe the process. |
Detects the presence and quantity of various DNA strands in a sample 1. Restriction endonucleases are used to cut high-molecular-weight DNA strands into smaller fragments.The DNA fragments are then electrophoresed on an agarose gel to separate them by size. 2. A sheet of nitrocellulose (or, alternatively, nylon) membrane is placed on top of (or below, depending on the direction of the transfer) the gel. Pressure is applied evenly to the gel (either using suction, or by placing a stack of paper towels and a weight on top of the membrane and gel), to ensure good and even contact between gel and membrane. 3. The membrane is then baked in a vacuum or regular oven at 80 °C for 2 hours (standard conditions; 4. The membrane is then exposed to a hybridization probe—a single DNA fragment with a specific sequence whose presence in the target DNA is to be determined. The probe DNA is labelled so that it can be detected, usually by incorporating radioactivity or tagging the molecule with a fluorescent or chromogenic dye. |
|
|
RNA transcription direction |
5' --> 3'. Transcribed off of template DNA, will match coding DNA strand (except T--> U) |
|
|
mRNA. What is poly vs monocistronic? |
Messenger RNA. Transcribed from template DNA strand via RNA polymerase in the nucleus of the cell. May undergo posttrascriptional modifications.
Monocistronic: One protein per mRNA. This is the case for eukaryotes
Polycistronic: Multiple proteins per mRNA. This is the case for prokaryotes. |
|
|
tRNA. Where is it found and what is charged tRNA? |
Transfer RNA. Has a 3-base pair anticodon sequence that pairs with RNA. Found in the cytoplasm and is charged when it has its corresponding amino acid attached to it.
Amino-acyl tRNA synthetase + two phosphate groups from ATP required to attach the amino group to the 3' end (CCA sequence) |
|
|
rRNA. Where is it found and what are ribozymes? |
Ribosomal RNA. Synthesized in the nucleolus and functions in the cytoplasm Ribozymes are ribosomal enzymes. |
|
|
Start and stop codons |
Start: AUG (MET) Stop: UAA, UGA, UAG / TAA, TGA, TAG |
|
|
Types of DNA coding mutations |
Point mutation: 1 base pair mutation -- Missense: changes the amino acid -- Silent : does not change the amino acid (degenerate) -- nonsense: changes to a premature stop codon Frameshift mutation: add/takes away nucleotides so reading frame is shifted. |
|
|
Promoter regions. What is a common promoter region for eukaryotes? |
Regions of the DNA where DNA transcription is initialized. In eukaryotes, this is often the TATA box (~ negative 25). RNA polymerase II is the main RNA polymerase for transcription. |
|
|
RNA Polymerase I, II, and III. Where is each located and what is the function? |
RNA Polymerase I: located in the nucleolus and synthesizes rRNA RNA Polymerase II: located in the nucleus and synthesizes hnRNA (heterogeneous nuclear mRNA) and snRNA (small nuclear RNA) RNA Polymerase III: located in the nucleus and synthesizes tRNA and some rRNA. |
|
|
Posttranscriptional modifications |
Splicing: spliceosomes excise introns from hnRNA via lariats. Alternative splicing allows for different variants of proteins from the same hnRNA. 5' methylguanine cap. Recognized by the ribosome as a binding site and also protects the mRNA from degradation in the cytoplasm 3' poly-A tail. Protects mRNA against rapid degradation in the cytoplasm. |
|
|
Spliceosomes. What are they made of? |
snRNA coupled to small nuclear ribonucleoproteins (SNRPs) snRNA/snRP complex recognizes splice sites for introns in posttranscriptional modification |
|
|
Transcription factors
|
Eukaryotic gene expression. Transcription-activating proteins that scan DNA for specific DNA-binding motifs. Usually have DNA-binding domain and an activation domain. Activation domain recruits several things, including other transcription factors and RNA polymerase. Transcription factor only results in low, baseline gene expression. |
|
|
Gene amplification |
Enhancers (binding to the regulator region). Usually further away from gene than promotor region, thus requires hairpin loop to connect to promotor region. Gene duplication: Gene may become duplicated many times in series, or use helicase and duplicate many times in parallel. |
|
|
Heterochromatin vs euchromatin
|
Heterochromatin is tightly condensed chromatin. Shows up dark in light microscopy. Euchromatin is less condensed, allowing for replication and transcription. Shows up light in light microscopy. |
|
|
What is used for histone remodeling
|
Histone acetylation by histone acetylases transition chromatin between heterochromatin and euchromatin. Acetylation works by acetylating lysine groups on histones, decreasing the positive charge and thus the interaction with DNA, opening the chromatin.
|
|
|
DNA methylation
|
DNA methylation silences genes. Usually occurs on cytosines and adenosines. Heterochromatin regions usually heavily methylated. Methylating DNA over time indicates the parent strand. During base excision repair, the parent strand is assumed to be the correct sequence and the daughter strand assumed to have the error. |
|
|
DNA response element
|
A sequence of DNA that binds only to specific transcription factors.
|
|
|
Difference between translation in prokaryotes and eukaryotes
|
Translation begins during transcription in prokaryotes. Translation and transcription are separated by location in eukaryotes.
|
|
|
Shine-Delgarno sequence
|
Region for binding of small ribosomal subunit (30S rRNA) in prokaryotes. Located in the 5' UTR. Initial amino acid is f-met
|
|
|
How to determine the start site for translation of mRNA in eukaryotes?
|
AUG codon. Always codes for methionine. (Also same in kingdom archae) |
|
|
Three stages of translation
|
Initiation (initiation factors) , elongation (elongation factors), and termination (releasing factors). All stages require GTP. |
|
|
Elongation
|
Use A, P, E sites. Aminoacyl tRNA site, collects charged tRNAs Peptidyl transfer site moves growing peptide chain from tRNA in P site to tRNA in A site, elongating the chain. Exit site. In termination, water is added to the polypeptide chain, allowing peptidyl transferase to hydrolyze amino acid chain from final tRNA. |
|
|
Posttranslational processing
|
Folding via chaperones Cleavage (into active forms) Subunits coming together for quaternary structure. Addition of biomolecules: 1. Phosphorylation: active / inactive 2. Carboxylation: often used as a calcium binding site. 3. Glycosylation: addition of oligosaccharides as proteins pass through ER and Golgi apparatus, often used to mark destination of protein 4. Prenylation: addition of lipids to certain membrane-bound enzymes. |
|
|
Operons. What is the structure and the model? What does it do.
|
Used in prokaryotic gene expression. Allows for transcription of a cluster of genes within a single mRNA (polycistronic). Jacob-Monod Model RPOS Regulator site, enhancers (accelerate expression) Promotor site, binding site for RNA polymerases) Operator site, binding site for repressors Structure Site, actual gene code. |
|
|
Two types of operons. Describe each and give an example
|
Inducible (Positive): Normally off because repressor is normally bound to operator site. Inducer proteins compete to bind to repressors, freeing the operator site and allowing expression. Example: lac operon. Lactose acts as inducer and allows for expression of lactase gene when lactose is in high abundance. Repressible (negative). Normally always off because repressor is inactive. Requires corepressor to cause the repressor to stop gene expression. Co-repressor is usually the end product, acting as negative feedback. Example is tryptophan operon, where binding two molecules of tryptophan to repressor shuts off the operon. |
|
|
Fluid mosaic model
|
Phospholipid bilayers. Allows nonpolar, small material through.
|
|
|
Lipid rafts
|
Part of a phospholipid bilayer that includes proteins and distinct signaling areas. Collection of similar lipids with or without associated proteins that serve as attachment points for other biomolecules. |
|
|
Glycoprotein coat
|
Carbohydrates associated with plasma membrane-bound proteins. High level of carbohydrates in cell walls of plants, bacteria, and fungi.
|
|
|
Flippase/floppase
|
Flips lipids/proteins/rafts in the phospholipid bilayer.
|
|
|
Chylomicrons
|
Located inside the small intestine. Transports triglycerides in.
|
|
|
Unsaturated fatty acids. Why are they important and give two examples of essential ones.
|
Increase membrane fluidity. Examples: alpha-linolenic acid and linoleic acid.
|
|
|
3 types of membrane proteins
|
Transmembrane - completely through Embedded - proteins found on the interior or exterior side Membrane - associated (peripheral) - proteins bound through electrostatic interactions with the lipid bilayer, especially at lipid rafts or to other transmembrane / embedded proteins. Integral protein - set consisting of transmembrane and embedded (the proteins connected to the membrane via partially hydrophobic domain) |
|
|
Types of lipids that can be found in cell membranes.
|
Phospholipids (bilayer) Steroids - increase fluidity Cholesterol - increase fluidity Triglycerides Free fatty acids Sphingolipids Waxes - more common in plants. |
|
|
What role does carbohydrates play in the cell membrane?
|
Generally located on the extracellular side. Act as signaling and recognition molecules. e.g. ABO blood types |
|
|
Cell adhesion molecules (CAMS)
|
Cell-cell junctions. Proteins that allow cells to recognize each other and contribute to proper cell differentiation and development. e.g. cadherin |
|
|
GAP junctions. What are they and what is another name for them?
|
Connexons, comprised of 6 connexins. Allow for direct cell-cell communications. Permits for movement of water and some solutes. Generally does not permit proteins. |
|
|
Tight junctions. What do they do and where are they found?
|
Watertight seal between cells to prevent solutes from going around cells via paracellular route. Forces solutes to go through cells to get to the other side. Found in the epithelial cells. Limit permeability enough to create transepithelial voltage difference. |
|
|
Desmosomes. What do they do and where are they found?
|
Bind adjacent cells by anchoring to their cytoskeletons. Found primarily between two layers of epithelial tissue. Hemidesmosomes bind epithelial cells to basement membranes. |
|
|
Types of passive intermembrane transport. Name and describe each. |
Simple diffusion: nonpolar or small molecules directly diffuse across cell membrane via concentration gradient. note: small & polar (but still uncharged) molecules may pass through. e.g. water
Facilitated diffusion:
Osmosis: Specific kind of simple diffusion involving water. |
|
|
What is a colligative property?
|
Properties of solutions that do not depend on the specific chemical properties of solutes, but rather just the concentrations.
|
|
|
Osmotic pressure formula
|
Osmetic pressure = iMRT, where i = van't Hoff factor (how many particles the molecule dissociates to in solution) M = molarity of solution R = ideal gas constant T = absolute temperature (in Kelvins) |
|
|
Facilitated diffusion
|
Simple diffusion for molecules normally impermeable. (too large, charged, or polar). Occurs via transmembrane integral proteins. E.g. channel or carrier protein. |
|
|
Channel vs carrier protein
|
Channel: like a hallway Carrier: like a revolving door. Occluded state is the moment of transition where carrier is closed on both sides of the bilayer. |
|
|
Active transport. Primary vs secondary |
Primary: consumes ATP to move molecules against gradient Secondary: uses the energy from one particle going down its gradient, to push a different particle up against its gradient. |
|
|
Symport vs antiport
|
Secondary active transport. In symport, both molecules moving same direction. In antiport, molecules move in opposite directions.
|
|
|
Pinocytosis
|
Endocytosis of fluids and dissolved particles.
|
|
|
Phagocytosis
|
Ingestion of large solids such as bacteria. (type of endocytosis)
|
|
|
Membrane potential (Vm)
|
Membrane potential across cell membrane. Resting potential is -40 to -80 mV in most cells.
|
|
|
Nernst Equation
|
E = (RT)/(ZF) ln [ion_out]/[ion_in] = 61.5/z * log [ion_out]/[ion_in] 61.5 assumes body temperature (310 K) |
|
|
Goldman-Hodgekin-Katz Voltage equation
|
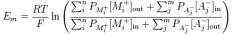
|
|
|
Na/K pumps. How many ions does it pump and in which direction?
|
3 sodium out for every 2 potassium in
|
|
|
Describe characteristics of the inner and outer mitochondrial membrane |
Inner membrane: many folds (cristae). Light levels of cardiolipin and no cholesterol. More restricted permeability Outer membrane: many large pores and highly permeable. |
|
|
Normal glucose concentration in peripheral blood. |
4-6mM |
|
|
GLUT 2 vs GLUT 4. Where are they located and what function do they serve? |
GLUT 2 is low affinity transporter in hepatocytes and pancreatic cells. In liver, captures excess glucose from portal vein after a meal to store for later use. In beta islet cells of the pancrease, acts as a glucose sensor for insulin release. GLUT 4 is in adipose tissue and muscle. Rate of glucose transport is elevated by insulin. (Insulin induces movement of GLUT 4 receptors to membrane). Km of GLUT 4 approximately equal to normal peripheral blood glucose level. |
|
|
Hexokinase vs glucokinase |
Phosphorylates glucose after transport by GLUT to trap in cells. Hexokinase found in most cells. Has low Hm, and is inhibited by glucose 6 phosphate Glucokinase is found in live cells and beta-islet pancreatic cells (with GLUT 2). High Km and is induced by insulin. |
|
|
Phosphofructokinase |
Rate controlling step / main controlling step for glycolysis PFK-1 inhibited by ATP and citrate, activated by AMP PFK-2 converts some Fructose 6 kinase to fructose 2,6 bisphosphate, which promotes PFK-1. Insulin promotes PFK-2 and glucagon inhibits it. PFK-2 exists in livers and overrides the inhibition on PFK-1 through fructose 2,6 bisphosphate control. |
|
|
List the rate limiting enzymes for the following processes: glycolysis fermentation glycogenesis glycogenolysis gluconeogenesis pentose phosphate pathway |
PFK-1 Lactate dehydrogenase Glycogen synthase glycogen phosphorylase fructose -1,6 bisphosphatase glucose-6-phosphate dehydrogenase |
|
|
Substrate level phosphorylation |
ADP to ATP using a high energy intermediate (e.g. 1,3 bisphosphoglycerate). Does not require oxygen. |
|
|
What activates pyruvate kinase? |
fructose 1,6 bisphosphate. Example of a feed-forward reaction. |
|
|
Fermentation |
Rate limiting enzyme is lactate dehydrogenase. Oxidation of NADH to NAD+ and reduction of pyruvate to lactate to regenerate NAD+ to allow glycolysis to continue. In yeast cells, pyruvate is reduced to ethanol and carbon dioxide. |
|
|
Irreversible enzymes in glycolysis |
Kinases: hexokinase, glucokinase, phosphofructokinase-1, pyruvate kinase |
|
|
High energy intermediates in glycolysis |
1,3 bisphosphoglycerate phosphoenolpyruvate (PEP) Each step generates 1 ATP. |
|
|
Net ATP yield of glycolysis |
2 ATP per glucose molecule |
|
|
2,3 BPG. Where is it located and what effect does it do? |
Biphosphoglycerate mutase converts 1,3 bisphosphoglycerate to 2,3 bisphosphoglycerate in erythrocytes.
This decreases affinity of red blood cells to oxygen, allowing for better unloading of oxygen in tissues. |
|
|
Important enzymes in galactose metabolism |
Galactokinase Galactose -1- phosphate uridyltransferase |
|
|
Important enzymes in fructose metabolism |
fructokinase aldolase B |
|
|
Pyruvate dehydrogenase. What does it do and what are the cofactors and coenzymes that make it up? |
Pyruvate + CoA --(NAD^+ --> NADH + CO2) --> Acetyl CoA Acetyl CoA and NADH has negative feedback on pyruvate dehydrogenase Cofactors include thiamine pyrophosphate, lipoid acid, CoA, FAD, NAD+. (TLC FADNAD) Coenzymes include: Pyruvate dehydrogenase (Mg2+ required also)Dihydrolipoyl tranacetylase, Dihydrolipoyl dehydrogenase, Pyruvate dehydrogenase kinase, Pyruvate dehydrogenase phosphatase. |
|
|
Glycogenin |
Core protein in glycogen granules. |
|
|
Glycogenesis. What is the rate limiting enzyme and its associated activators/inhibitors? |
Glycogen synthase. Activated by insulin and glucose 6-phosphate. Inhibited by epinephrine and glucagon. |
|
|
Glycogenolysis. What is the rate limiting enzyme and its associated factors? |
Glycogen phosphorylase. Activated by glucagon, AMP, and epinephrine. Inhibited by ATP Breaks alpha 1,4 glycosidic linkages in glucogen. |
|
|
Glycogen debranching enzyme steps |
1. Glucosyl transferase hydrolyzes the alpha 1,4 nearest the branch point. 2. Transfer the just hydrolyzed chain to end of another branch. 3.. Glycosidase cuts of the glucose 1,6 bond, releasing a free glucose. |
|
|
Gluconeogenesis. Where does it occur? What activates/inhibits this pathway? What are the substrates? What are the important intermediates? |
Liver (and to a smaller extent kidneys). Promoted by glucagon and epinephrine. Inhibited by insulin Substrates: Glycerol - 3 - phosphate, lactate, glucogenic amino acids (all except lysine and leucine) Important intermediates: lactate, alanine, glycerol 3-phosphate |
|
|
Important enzymes of gluconeogenesis. What activates/inhibits each of them? |
Pyruvate carboxylase (found in the mitochondrion) + malate/aspartate shuttle). Part 1 of bypassing pyruvate kinase. Activated by acetyl CoA. Phosphoenolpyruvate carboxykinase (PEPCK). Step 2 of bypassing acetyl CoA. OAA (+ GTP) --> PEP (+ GDP). Induced by glucagon and cortisol. Fructose - 1,6 - biphosphatase. Rate limiting step of gluconeogenesis. Activated by ATP and inhibited by fructose 2,6 bisphosphate and AMP. Bypasses PFK-1 Glucose 6 phosphatase. Found only in the lumen of the ER in liver cells. Glucose 6 phosphate transported to ER and free glucose diffuses out, where it can be released via the GLUT transporters. Circumvents hexokinase/glucokinase. |
|
|
Cori cycle |
Glucose converted to lactic acid in red blood cells via anaerobic glycolysis. Lactate is transported to liver, where it reforms glucose via gluconeogenesis. |
|
|
Pathways to generating Acetyl-CoA (5)
|
Glycolysis Fatty acid (beta) oxidation Amino acid catabolism Ketones Alcohol |
|
|
TCA Cycle. What is the net equation? Mneumonic?
|
Pyruvate, 2x NAD+, FAD, GDP/Pi to 2x CO2, 2x NADH, FADH2, GTP. Please can I keep selling seashells for money officer? |
|
|
TCA Step 1. Citrate formation. Reactants, products, enzyme.
|
Oxaloacetate + Acetyl CoA + H2O --citrate synthase--> Citrate + H+ (Condensation Reaction) |
|
|
TCA Step 2. Citrate isomerization
|
Citrate --aconitase--> Isocitrate |
|
|
TCA Step 3. alpha-ketoglutarate and CO2 formation.
|
Rate limiting enzyme Isocitrate + NAD+ --isocitrate dehydrogenase --> alpha-ketoglutarate + CO2 + NADH |
|
|
TCA Step 4. Succinyl-CoA and CO2 formation
|
alpha-ketoglutarate + CoA + NAD+ --alpha-ketoglutarate dehydrogenase complex--> Succinyl-CoA + NADH + CO2 |
|
|
TCA Step 5. Succinate formation
|
Succinyl CoA + GDP/Pi -- succinyl-CoA synthetase --> succinate + GTP |
|
|
nucleosidediphosphate kinase
|
Used in the TCA cycle. Generates 1 ATP from 1 GTP. This is the only ATP directly created from the TCA cycle.
|
|
|
TCA step 6 Fumarate formation
|
Occurs on the inner membrane. Succinate + FAD --succinate dehydrogenase (a flavoprotein) --> Fumarate + FADH2 |
|
|
Electron transport chain. ATP per NADH equivalence. ATP per FADH2 equivalence.
|
1 NADH = 2.5 ATP 1 FADH2 = 1.5 ATP |
|
|
TCA Step 7 Malate formation
|
Fumarate --Fumarase--> L-Malate |
|
|
TCA Step 8 OAA formation
|
L-Malate + NAD+ --malate dehydrogenase--> OAA + NADH |
|
|
Glucose ATP yield breakdown.
|
Glycolysis (per glucose): 2 ATP 2 NADH Pyruvate dehydrogenation (per pyruvate): 1 NADH TCA Cycle (per pyruvate): 3 NADH 1 FADH2 1 GTP Total yield per pyruvate: 4 NADH --> 10 ATP 1 FADH2 --> 1.5 ATP 1 GTP --> 1 ATP 12.5 ATP per pyruvate 25 ATP per glucose from PDH and TCA cycle. + 2 ATP + 2 NADH (~5 Glucose) from glycolysis Total yield : 30-32 ATP per glucose |
|
|
Pyruvate dehydrogenase regulation
|
Inhibition: Acetyl-CoA, Phosphorylation by PDH kinase during periods of high ATP Activation: Dephosphorylation by PDH phosphatase during periods of high ADP. |
|
|
TCA cycle regulation. What are the key regulatory enzymes? What is the rate-limiting enzyme?
|
Rate limiting enzyme is isocitrate dehydrogenase. Key enzymes are Citrate synthase, isocitrate dehydrogenase, alpha-ketoglutarate dehydrogenase complex |
|
|
Citrate synthase regulators (4)
|
Allosteric inhibitors: ATP, NADH, Citrate, succinyl-CoA |
|
|
Isocitrate dehydrogenase regulators (2)
|
Inhibitors: ATP, NADH Activators: ADP, NAD+ |
|
|
Alpha-ketoglutarate dehydrogenase complex regulators
|
Inhibitors: succinyl-CoA, NADH, ATP Activators: ADP, Calcium ions. |
|
|
ETC Complex I.
|
Electron goes from NADH to FMNH2 to Fe-S to Coenzyme Q. Moves 4 H+ into intermembrane space. |
|
|
ETC Complex II
|
Electron goes from Succinate to FADH2 to Fe-S to Coenzyme Q. No protons are moved in this process. |
|
|
ETC Complex III
|
Electron moves from Coenzyme Q to 2x Cytochrome C. Q Cycle 4 protons are moved to the intermembrane space in this process. |
|
|
ETC Complex IV
|
Electrons move from Cytochrome C to oxygen and form water. 2 Protons are moved across the membrane in this process. |
|
|
NADH Shuttles. Two types and which is more efficient?
|
Glycerol 3 phosphate shuttle: glycerol-3 dehydrogenase oxidizes NADH in cytosol and reduces FAD in inner membrane. Malate-aspartate shuttle: Malate dehydrogenase reduces oxaloacetate into malate, which can cross the membrane. Malate reduced back into oxaloacetate, generating a NADH inside. Oxaloacetate goes back out as aspartate. |
|
|
ATP Synthase. Components of the enzyme and where is it located?
|
Spans inner membrane and protrudes into the matrix. F0 spans the matrix, functions as ion channel. F1 utilizes energy from electrochemical gradient to phosphorylate ADP into ATP. |
|
|
Regulators of TCA and ETC.
|
NADH accumulation is inhibitory. ADP accumulation is activating. |
|
|
Digestion of lipids (Emulsification and hydrolysis, where and how?)
|
Minimal digestion in mouth and stomach. Emulsification in the duodenum via bile salts. Hydrolysis via pancreatic enzymes: pancreatic lipase, colipase, and cholesterol esterase in the small intestine. After hydrolysis: 2-monoacylglycerol |
|
|
Absorption of lipids
|
Micelle formation of: 2-monoacylglycerol Free fatty acids Cholesterol Bile salts Micelle diffusion to the brush border. Re-esterified and packaged into chylomicrons, which leave via lacteals to the lymphatic system and enter the bloodstream via thoracic duct. Small FFAs diffuse into bloodstream directly |
|
|
Lacteals
|
Vessels of the lymphatic system
|
|
|
Chylomicrons
|
Lipid transporters
|
|
|
Thoracic duct
|
Lymphatic to bloodstream duct.
|
|
|
HSL. Where is it? What is it? What does it do? What activates it?
|
Low insulin levels activate hormone-sensitive lipase in adipose tissue. It hydrolyzes triglycerides into free fatty acids and glycerol. It is activated by low insulin, epinephrine, and cortisol.
|
|
|
LPL. What is it, what does it do?
|
Lipoprotein Lipase. Necessary for metabolism of chylomicrons and VLDL (very low density lipoproteins).
|
|
|
Lipoproteins. What are they and what are the types?
|
Lipid carriers for triacylglycerol and cholesterol. Least dense Chylomicron VLDL IDL LDL HDL Most Dense |
|
|
Chylomicron function.
|
Transports dietary triacylglycerols and cholesterol from intestine to tissues.
|
|
|
VLDL Function
|
Very low density lipoproteins. Transports triacylglycerols from liver to tissues |
|
|
IDL Function
|
Intermediate-density lipoproteins Remnant of VLDL after triacylglycerol is removed. Transition between VLDL and LDL (triacylglycerol vs cholesterol) |
|
|
LDL Function.
|
Low density lipoprotein. (Bad Cholesterol) Majority of blood cholesterol associated with LDL. Functions to transport cholesterol to tissues. |
|
|
HDL Function. Where is it synthesized?
|
High density lipoprotein. (Good cholesterol) Synthesized in the liver and intestines. Cleans up excess cholesterol in blood and sends to excretion. |
|
|
Where may cholesterol be synthesized? What are the reagents? What is the rate-limiting enzyme?
|
Liver. Driven by Acetyl-CoA, ATP, NADPH. Rate limiting enzyme: HMG CoA reductase (mevalonic acid synthesis) |
|
|
Cholesterol synthesis regulators.
|
Negative feedback by cholesterol product. Activated by insulin. Regulation of HMG CoA reductase expression from genome. |
|
|
LCAT. Name, location, function, and activators.
|
Lecithin-cholesterol acyltransferase. Found in the bloodstream. Adds fatty acid to cholesterol to form cholesterol esters. Activated by HDL apoproteins. |
|
|
CETP. Name, location, function. |
Cholesteryl ester transfer protein. Located in blood cells. Facilitates transfer of cholesterol esters (made by LCAT) from HDL to IDL, forming LDL. |
|
|
Apoproteins
|
Protein component of lipoproteins.
|
|
|
HMG CoA reductase. Function, location, product.
|
Found in SER. Rate limiting step of cholesterol synthesis. Product is mevalonic acid.
|
|
|
Nontemplate synthesis. What is it? Give two examples.
|
Biosynthesis without DNA. E.g. lipid and cholesterol synthesis.
|
|
|
Fatty acid biosynthesis. Where does it occur? What are the key enzymes and what are they stimulated by? What is the primary end product?
|
Occurs in the liver (and to a smaller extent in the adipose tissue), products transferred to adipose tissue. Key enzymes are acetyl-CoA carboxylase and fatty acid synthase. Stimulated by insulin. Primary end product is palmitic acid (palmitate) |
|
|
Citrate lyase. What does it do and where?
|
Citrate diffuses out of matrix into the cytosol via citrate shuttles after large meal (Accumulation of citrate in the matrix). Citrate lyase breaks it up into OAA and Acetyl-CoA again.
|
|
|
What is the rate limiting enzyme of fatty acid synthesis?
|
Acetyl-CoA Carboxylase.
|
|
|
Acetyl-CoA Carboxylase. What are the coenzymes/cofactors? What does it do? What activates it?
|
Requires biotin and ATP. Adds CO2 to acetyl-CoA to form malonyl-CoA. Activated by insulin and citrate.
|
|
|
Fatty Acid Synthesis. Net equation. |
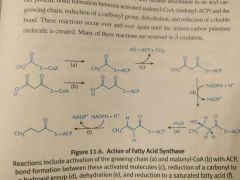
7 malonyl-CoA + 1 acetyl-CoA + 14 NADPH --> Palmitate + 7 CO2 + 14 NADP Attachment to ACP Binding to growing chain. Carboxyl reduction Dehydration Reduction of double bond. |
|
|
Fatty acid catabolism. How and where does it occur? What stimulates and inhibits this process?
|
Occurs in the mitochondria via beta-oxidation. Insulin inhibits this while glucagon stimulates it.
|

