![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
78 Cards in this Set
- Front
- Back
|
What are the two types of nucleic acids? |
* RNA - Genetic material for viruses - Functions as adapter, structural and catalytic molecule - Functions as messenger * DNA - Most common genetic material
|
|
|
Bacteriophage phi 174 nucleotide number? |
5386 nucleotides |
|
|
Bacteriophage lambda base pair no.? |
48,502 base pairs |
|
|
Escheria coli base pair number? |
4.6 x 10^6 base pairs |
|
|
Human DNA (haploid) base pair number ? |
3.3 x 10^9 base pairs |
|
|
What are components of a polynucleotide chain? |
1. Nitrogenous Base - Purine (Adenine, Guanine) - Pyrimidine ( Thymine, Cytosine, Uracil) 2. Penrose sugar ( Ribose in RNA and Deoxyribose in DNA) 3. Phosphate group |
|
|
Nucleoside |
Nitrogenous Base linked to pentode sugar through N-glycosidic linkage |
|
|
Nucleotide |
Phosphate group is linked to 5'-OH of a nucleoside through phoshpoester linkage
Linkage between two nucleotides is phosphodiester |
|
|
What is 5' end ? |
End of the polynucleotide chain with a free OH at 5' end of ribose sugar. |
|
|
What is 3' end? |
End of the polynucleotide chain with a free OH at 3' end of ribose sugar. |
|
|
Who was the first one to identify DNA? |
- Friedrich Meischer in 1869. - Considered it an acidic substance present in the nucleus and called nuclein . |
|
|
Who proposed the Double Helix Model? |
- 1953, James Watson and Francis Crick on the basis of X-ray diffraction data from Maurice Wilkins and Rosalind Franklin proposed DH model. |
|
|
What was the base pairing proposition given by Erwin Chargaff? |
For a double stranded DNA, the ratios between Adenine and Thymine , Guanine and Cytosine are constant and equal to one. |
|
|
What unique properties did base pairing of DNA confer? |
1. The opposing strands are complementary and if sequence of one is known, the other can be predicted. 2. If each strand of DNA acts as template for synthesis, the daughter DNA would be identical to the parental DNA molecule. |
|
|
What are the "Salient Features" of double helix model? |
1. Consists of two polynucleotide chains with a backbone made of sugar-phosphate groups and the nitrogenous bases project inwards. 2. The two strands have anti parallel polarity ( if one is 5' - 3', the other is 3' - 5') and are coiled in a right hand fashion. 3. Complementary base pairing occurs through hydrogen bonds. "A" pairs with "T" through 2 H bonds and "G" pairs with "C" through 3 H bonds. A pyrimidine always pairs with a purine, this ensures uniform distance between the two. 4. - Distance between two base pairs is 0.34 nm - Complete turn contains 10 base pairs - Pitch of the helix is 0.34 nm 5. Plane of one base pair stacks over the other in a double helix which confers additional stability. |
|
|
Central Dogma? |
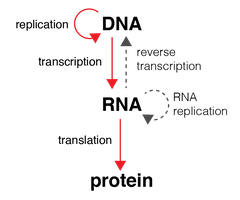
Proposed by Francis Crick |
|
|
Length of DNA? |
- Length = no. of bp x dist. between bp - Human DNA = 6.6 x 10^9 x 0.34 x 10^-9 = 2.2 m |
|
|
DNA packaging in prokaryotes? |
Nucleoid in prokaryotes, where negatively charged DNA is held within positively charged non-histone basic proteins. |
|
|
DNA packaging in eukaryotes discovery? |
- Roger Kornberg 1974, reported that chromosome is made up of DNA and protein - Tatum and Beadle reported that chromatin fibre looks like beads on string and beads are repeated units of proteins. |
|
|
Proteins associated with DNA packaging in eukaryotes? |
- Basic proteins - Histones - Acidic proteins - Non-Histone Chromosomal Proteins |
|
|
DNA Packaging in eukaryotes? |
- Histones ( +vely charged basic protiens ) are organised into Histone Octamer (unit of 8 molecules) - Histone Octamer consists of 4 protiens in pairs (H2A, H2B, H3, H4) - -vely charged DNA is wound around the Histone Octamer to form a Nucleosome (Consists of 200 bp) - Repeating units of nucleosomes form chromatin - Chromatin is packaged to form chromatin fibre - During metaphase, chromatin fibre condenses into chromosomes - Packaging of chromatin at higher level requires NHC protiens |
|
|
Difference between euchromatin and heterochromatin? |
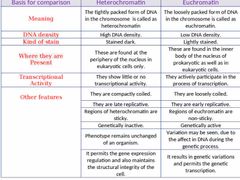
|
|
|
How does protein acquire charge? |
They acquire charge depending on abundance of amino acid residues within charged side chains |
|
|
What basic amino acids do histones contain? |
Lysine and Arginine |
|
|
What were the three experiments conducted in the search for genetic material? |
1. Transforming Principle - Frederick Griffith 1928 2. Biochemical nature of transforming principle - Oswald Avery, Colin MacLeod and Maclyn McCarty 1933-44 3. Proof for DNA as Genetic Material - Alfred Hershey and Martha Chase 1952 |
|
|
Transforming principle experiment |
Experiment - Conducted by Frederick Griffith in 1928 with streptococcus pneumoniae - Observed two strains of bacteria, Smooth S-strain (Virulent polysaccharide mucous coat) & Rough R-strain lacked the mucous coat - When live S-strain injected, mice died while with live R-strain, the mice lived - When heat killed S-strain injected mice lived, however when heat killed S-strain mixed with live R-strain the mice died Conclusion 1. R-strain had been "transformed" by heat killed S-strain bacteria 2. Some "transforming principle" transferred from heat-killed S-strain enabled R-strain to develop a polysaccharide coat and become virulent 3. He assumed this was due to transfer of genetic material but biochemical nature of genetic material wasn't defined |
|
|
Biochemical Nature of Transforming Principle |
- Oswald Avery, Colin MacLeod and Maclyn McArty, from 1933-44 worked to determine biochem. nature of transforming principle. - They purified the biochemicals (Proteins, DNA & RNA) from heat-killed S-strain to see which ones could transform live R-strain to S-strain - They observed that DNA alone could cause the transformation - Moreover, they discovered that RNases and Proteases didn't inhibit the transformation whereas DNase did inhibit the process - Therefore DNA was genetic material |
|
|
Proof that DNA was the Genetic Material |
- Alfred Hershey and Martha Chase in 1952 worked with viruses called bacteriophages. The bacteriophage attaches to the bacteria and its genetic material enters the bacteria. The bacteria treats this viral DNA as its own and manufactures more viral cells. Procedure - Bacteriophages were grown in a medium containing radioactive Phosphorous (P32) and some in radioactive Sulphure ( S35) - Viruses grown in P32 had radioactive DNA ( As DNA contains phosphorous ) - While viruses grown in S35 had radioactive proteins ( As proteins contain sulphur ) - Infection Radioactive phages were allowed to attach to E.coli - Blending After infection, viral coats were removed by agitating in a blender - Centrifugation Viral particles were separated from the bacteria by spinning in a centrifuge Observation & Conclusion - Only radioactive P32 was found to be associated with the bacterial cell, whereas S35 was found in the surrounding material - Therefore, protein coat didn't enter the bacteria and only DNA entered - Therefore, DNA was the genetic material
|
|
|
What criteria should genetic material fulfil? |
1. Should be able to undergo replication 2. Should be chemically and structurally stable 3. Should provide scope for mutations required for evolution 4. Should be able to express itself in terms of mendelian chracters |
|
|
Why is DNA more stable than RNA? |
DNA - Double stranded - Strands if separated come back together by heating - Thymine ( 5 methyl Uracil ) is more stable than uracil RNA - 2'-OH groups on every nucleotide make it labile, reactive and easily degradable |
|
|
Difference between RNA and DNA |
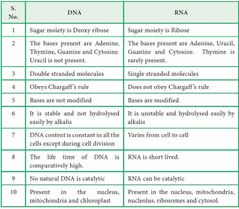
|
|
|
What does "semi-conservative" imply? |
Watson and Crick proposed that each strand of double stranded DNA separated and acted as template for synthesising complimentary strand After completion of replication, each DNA had one parental and one newly synthesised strand |
|
|
Experimental proof for semi-conservative DNA replication |
- Matthew Meselson and Frankiln Stahl in 1958 performed experiments on E. coli to prove that DNA replication is semi-conservative Procedure - They grew E. coli in a medium containing 15 NH4Cl (15N is a heavy isotope of nitrogen) for several generations as the only nitrogen source. As a result 15N got incorporated into the DNA - Heavy DNA can be differentiated from normal DNA by centrifugation in a CsCl (Cesium Chloride) density gradient - They transferred the E. coli to a medium containing normal 14NH4Cl and took sample at definite intervals as cells multiplied - Extracted DNAs were centrifuged and measured to get their densities. DNA extracted after one generation ( 20 minutes) showed intermediate hybrid density - DNA extracted after two generations (40 minutes) showed equal amounts of hybrid and light DNA - After 80 minutes light DNA increased while hybrid DNA maintained itself |
|
|
What experiment was Taylor involved in? |
Taylor and his colleagues in 1958 experimented with Vicia faba using radioactive thymidine to detect newly synthesised DNA and proved that DNA replicates semiconservatively even in chromosomes |
|
|
What are dNTPs? |
Deoxyribonucleoside triphosphates act on substrates and provide energy for polymerisation Terminal phosphates in dNTPs are high energy phosphates same as ATP |
|
|
DNA Polymerase I |
Involved in exonuclease, removes RNA and replaces it with newly synthesised DNA |
|
|
DNA Polymerase III (DNA dependent DNA polymerase) |
Is the main enzyme, adds nucleotides in 5' - 3' direction Uses DNA as template to catalyse polymerisation of deoxyribonucleotides |
|
|
Helicase |
Opens helix by breaking hydrogen bonds between nitrogen bases |
|
|
Ligase |
Seals the gap between Okazaki fragments to create a continuous DNA strand |
|
|
Primase |
Synthesises RNA primers to start replication |
|
|
Single Stranded Binding Proteins |
Bind to single stranded DNA to prevent Hydrogen bonding between the two strands |
|
|
Topoisomerase III (DNA gyrase) |
Relaxes supercoiled DNA and hence making it more accessible for initiation of replication |
|
|
Why does DNA polymerase only add nucleotides in 5'-3' direction? |
Because it needs a free 3'-OH group to which it can add nucleotides by forming a phosphodiester bond between 3'OH and 5' phosphate of next nucleotide |
|
|
Replication (Initiation) |
- Occurs at specific nucleotide sequence called origin of replication or "ori" - Topoisomerase III (DNA gyrase) relaxes the supercoiled DNA - Helicase breaks the hydrogen bonds between nitrogenous bases and separates the two strands - DNA near each replication fork is coated with Single Stranded Binding Proteins which prevent the DNA from rewinding into a double helix - Unwinding of DNA forms a Y-shaped replication fork |
|
|
Replication (Elongation) |
- Primase initiates replication of DNA strand oriented 3'-5' and results in the formation of 10-60 nucleotide long primer RNA - The free 3'-OH of RNA primer provides initiation point for DNA polymerase - DNA polymerase progressively adds nucleotides on the 3'-5' direction such that replication is continuous - 3'-5' leading strand; 5'-3' lagging strand - Replication of lagging strand occurs by formation of small fragments called Okazaki fragments that are later joined by action of DNA ligase |
|
|
Difference between transcription and replication |

|
|
|
Why are both strands not copied during transcription? |
- If both act as template they would code for RNA with different sequences which in turn would code for proteins with different sequence of amino acids. As a result, one strand of DNA would code for two different proteins which would further complicate the genetic machinery - If two complementary strands of RNA are produced simultaneously, they would coil into double stranded RNA which wouldn't allow RNA to be translated into proteins |
|
|
What is a transcription unit composed of? |
It consists of : 1. Promoter - Binding site for RNA polymerase for initiation of transcription 2. Structural gene - Codes for enzyme/protein 3. Terminator - Region where transcription ends |
|
|
What is the template strand in transcription? |
The RNA polymerase catalyses reaction in 5'-3' direction, so 3'-5' polarity strand acts as template strand |
|
|
What is coding strand? |
The strand with 5'-3' polarity doesn't code for any RNA but has the same sequence as RNA ( with the exception of thymine in place of uracil) and all references are made along the coding strand |
|
|
Where is promoter located? |
It is located at the 5' end of structural gene |
|
|
What is a gene? |
Sequence of DNA coding for RNA, functional unit of inheritance |
|
|
What is cistron? |
A segment of DNA coding for a polypeptide |
|
|
What is monocistronic? |
In eukaryotes transcriptional unit possesses a structural gene that codes for only one polypeptide chain |
|
|
What is polycistronic? |
In prokaryotes, transcriptional unit possesses a structural gene that codes for many polypeptides |
|
|
What are exons? |
Coding or expressed sequences of DNA that appear in mature/processed RNA |
|
|
What are introns? |
Intervening sequences of DNA that do not appear in mature or processed RNA |
|
|
What are the types of RNAs? |
- tRNA: Brings amino acids and reads genetic code - rRNA: Plays structural and catalytic role during synthesis of proteins - mRNA: Provides template for protein synthesis |
|
|
Transcription in bacteria |
1. Initiation - RNA polymerase binds to promoter and initiates transcription - Uses NTP(nucleoside triphosphate) as substrate and and polymerises in a template dependent fashion following rules of complementarity - Sigma factor recognises the start signal and promoter region on DNA 2. Elongation - RNA polymerase after initiation loses sigma factor and continues the polymerisation of ribonucleotides to form RNA. It also facilitates the opening of the helix 3. Terminantion - Once RNA reaches the terminator region, the RNA polymerase is separated from RNA-DNA hybrid. As a result nascent RNA separates. This process is called termination and is facilitated by rho; termination factor |
|
|
How does RNA polymerase catalyse all three steps? |
- It can actually only catalyse the elongation phase but it catalyses initiation and termination by associating transiently with sigma and rho factor - They alter the specificity of RNA polymerase hence allowing it to initiate or terminate |
|
|
How are transcription and translation coupled in bacteria? |
- mRNA doesn't require any processing to become transcriptionally active - transcription and translation take place in the same compartment because there is no separation between cytosol and nucleus in prokaryotes - As a result translation begins even before mRNA is fully transcribed. |
|
|
What are the types of RNA polymerase found in eukaryotes? |
- RNA polymerase I - Transcribes rRNAs (28S, 18S & 5.8S) - RNA polymerase II - Transcribes precursor of mRNA; heterogenous RNA (hnRNA) - RNA polymerase III - Transcribes tRNA, 5S rRNA & snRNA (small nuclear RNA) |
|
|
What are the additional steps in transcription for eukaryotes? |
- In eukaryotes, the primary transcripts contain exons as well as introns, and introns do not code for polypeptides, as a result it undergoes these additional steps 1. Splicing - Introns are removes from hnRNA and exons are joined in a defined order 2. Capping - Methyl guanosine triphosphate is added to the 5'-end of hnRNA 3. Tailing - Adenylate residues (200-300) are added to 3'-end in a template independent manner Now fully processed hnRNA is transformed into mRNA and ready for translation |
|
|
What is the purpose of capping and tailing? |
- Methyl guanosine triphosphate *Protects the transcript from being broken down *Helps ribosome to attach to mRNA and aids in translation - Polyadenylate tail *Makes transcript more stable *Helps it to get exported from nucleus to cytosol |
|
|
What is Genetic Code? |
Relation between sequence of nucleotides in mRNA and sequence of amino acids in polypeptides |
|
|
What is a codon? |
A sequence of three nucleotides which together form a unit of genetic code in a DNA or RNA molecule. |
|
|
George Gamow |
- Physicist who proposed that since there are 4 bases which code for 20 amino acids, in order for bases to be able to code for all the amino acids, code should be made up of 3 nucleotides therefore, 64 codons code for 20 amino acids |
|
|
Har Gobind Khorana |
Developed chemical method for synthesising RNA molecules with defined base combinations to develop the genetic code |
|
|
Marshall Nirenburg |
Developed cell-free system for protein synthesis and artificially synthesised proteins to understand nature of codon |
|
|
Severo Ochoa |
Demonstrated that polynucleotide phosphorylase also helped in polymerising RNA with defined sequences in a template-independent manner |
|
|
Salient features of genetic code |
1. The codons are a triplet, out of 64 codons only 61 code for 20 amino acids the other three codons (UAG, UAA, UGA) do not code for anything and are stop codons 2. One codon only codes for one amino acid, hence the code is unambiguous and specific 3. Some amino acids are coded for by more than one codon, hence the code is degenerate 4. The code is read on mRNA in a contiguous fashion, there are no punctuations 5. The genetic code is universal, a particular codon codes for the same amino acid in all organisms, from humans to bacteria with the exception of some mitochondria ans protozoans 6. (AUG) has dual functions it codes for Methionine and acts as start codon |
|
|
What is a mutation? |
Sudden inheritable change in the genetic material |
|
|
Point mutation |
Change in single base pair which is replaced by another base pair In case of sickle cell anaemia Point mutation in beta globin chain results in change of glutamate to valine |
|
|
Frameshift mutation |
Change of reading frame due to insertion or deletion of base pairs -Insertion : Addition of one or more nucleotides to a segment of DNA. Insertion in x3 doesn't change reading frame but adds a new amino acid -Deletion : Removal of one or more base pairs from a segment of DNA. Deletion in x3 doesn't change reading frame but removes an amino acid. |
|
|
tRNA structure |
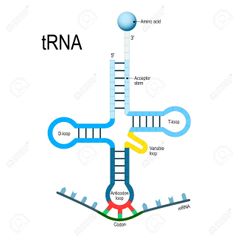
- Secondary structure is clover leaf like while tertiary structure is inverted L shaped - tRNA has 5 arms/loops 1. Anticodon loop, which has bases complementary to code 2. Amino acid acceptor end, where amino acid binds 3. T loop, which helps in binding to ribosome 4. D loop, which helps in binding to aminoacyl synthetase 5. Variable arm - There is no tRNA for stop codons |
|
|
What is translation? |
Process of synthesis of protein from amino acids, sequence and order of amino acids being defined by the sequence and order of bases in mRNA. Amino acids are joined by peptide bond |
|
|
What is a translational unit compose of? |
- Consists of mRNA sequence from 5'-3' - Start codon - Region coding for amino acids - Stop codons - UTR (Untranslated regions) ensure efficient translation process |
|
|
Translation (Initiation) |
- Activation of amino acids |

